mod.powers.train <- lm(y ~ poly(x, degrees = 25), data = training_xy)
Metrics::rmse(training_xy$y, predict(mod.powers.train))[1] 987.8615[1] 1002.229| OLS | |
|---|---|
| (Intercept) | -17.447 |
| (31.640) | |
| x | 7.676 |
| (0.543) | |
| Num.Obs. | 1000 |
| R2 | 0.167 |
| R2 Adj. | 0.166 |
| AIC | 16656.8 |
| BIC | 16671.6 |
| Log.Lik. | -8325.414 |
| F | 200.071 |
| RMSE | 998.72 |
[1] "Signal vs. Noise" x y
Min. :-99.919 Min. :-3495.10
1st Qu.:-53.554 1st Qu.: -786.51
Median : -3.720 Median : -60.38
Mean : -1.075 Mean : -49.86
3rd Qu.: 51.472 3rd Qu.: 722.85
Max. : 99.698 Max. : 3155.77 x y
Min. :-99.952 Min. :-3410.78
1st Qu.:-55.411 1st Qu.: -546.54
Median : -5.878 Median : 58.90
Mean : -7.446 Mean : 22.05
3rd Qu.: 42.628 3rd Qu.: 702.45
Max. : 98.159 Max. : 3910.81 XGBoost
library(xgboost))library(tidymodels)
island, sex
penguins <- drop_na(penguins)
penguin_dummies <- recipe(species ~ ., penguins) |>
step_dummy(all_nominal_predictors(), one_hot = TRUE)
# see ?step_dummy for other encoding options
head(penguins)# A tibble: 6 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen 36.7 19.3 193 3450
5 Adelie Torgersen 39.3 20.6 190 3650
6 Adelie Torgersen 38.9 17.8 181 3625
# ℹ 2 more variables: sex <fct>, year <int>workflow that combines model and data preprocessing## Model & data preparation
species_pred_wf <- workflow() |>
add_model(mod.boost) |>
add_recipe(penguin_dummies)
## Cross-validation
specied_pred_cv <- vfold_cv(penguins, v = 5)
## Learning rate tuning
lr_tuning <- tune_grid(
species_pred_wf,
resamples = specied_pred_cv,
grid = random_lr,
metrics = metric_set(bal_accuracy),
control = control_grid(verbose = FALSE)
)
lr_tuning# Tuning results
# 5-fold cross-validation
# A tibble: 5 × 4
splits id .metrics .notes
<list> <chr> <list> <list>
1 <split [266/67]> Fold1 <tibble [10 × 5]> <tibble [0 × 3]>
2 <split [266/67]> Fold2 <tibble [10 × 5]> <tibble [0 × 3]>
3 <split [266/67]> Fold3 <tibble [10 × 5]> <tibble [0 × 3]>
4 <split [267/66]> Fold4 <tibble [10 × 5]> <tibble [0 × 3]>
5 <split [267/66]> Fold5 <tibble [10 × 5]> <tibble [0 × 3]># A tibble: 10 × 3
learn_rate mean std_err
<dbl> <dbl> <dbl>
1 0.186 0.989 0.00420
2 0.176 0.989 0.00420
3 0.230 0.989 0.00420
4 0.0449 0.986 0.00510
5 0.00461 0.983 0.00504
6 0.00852 0.983 0.00504
7 0.00319 0.983 0.00504
8 0.00143 0.983 0.00504
9 0.0270 0.980 0.00494
10 0.0374 0.980 0.00494## Select best learning rate
best_lr <- lr_tuning |> select_best()
## Set best learning rate for the model
species_pred_wf_best1 <- species_pred_wf |>
finalize_workflow(best_lr)
species_pred_wf_best1══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: boost_tree()
── Preprocessor ────────────────────────────────────────────────────────────────
1 Recipe Step
• step_dummy()
── Model ───────────────────────────────────────────────────────────────────────
Boosted Tree Model Specification (classification)
Main Arguments:
learn_rate = 0.186431577754487
Computational engine: xgboost library(colorspace)
set.seed(1)
penguin_fit <- species_pred_wf_best1 |>
fit(penguins)
penguins$pred <- penguin_fit |>
predict(penguins) |>
pull(.pred_class)
penguins |>
mutate(pred = as.factor(pred)) |>
conf_mat(species, pred) |>
autoplot(type = 'heatmap') +
scale_fill_continuous_diverging("Purple-Green")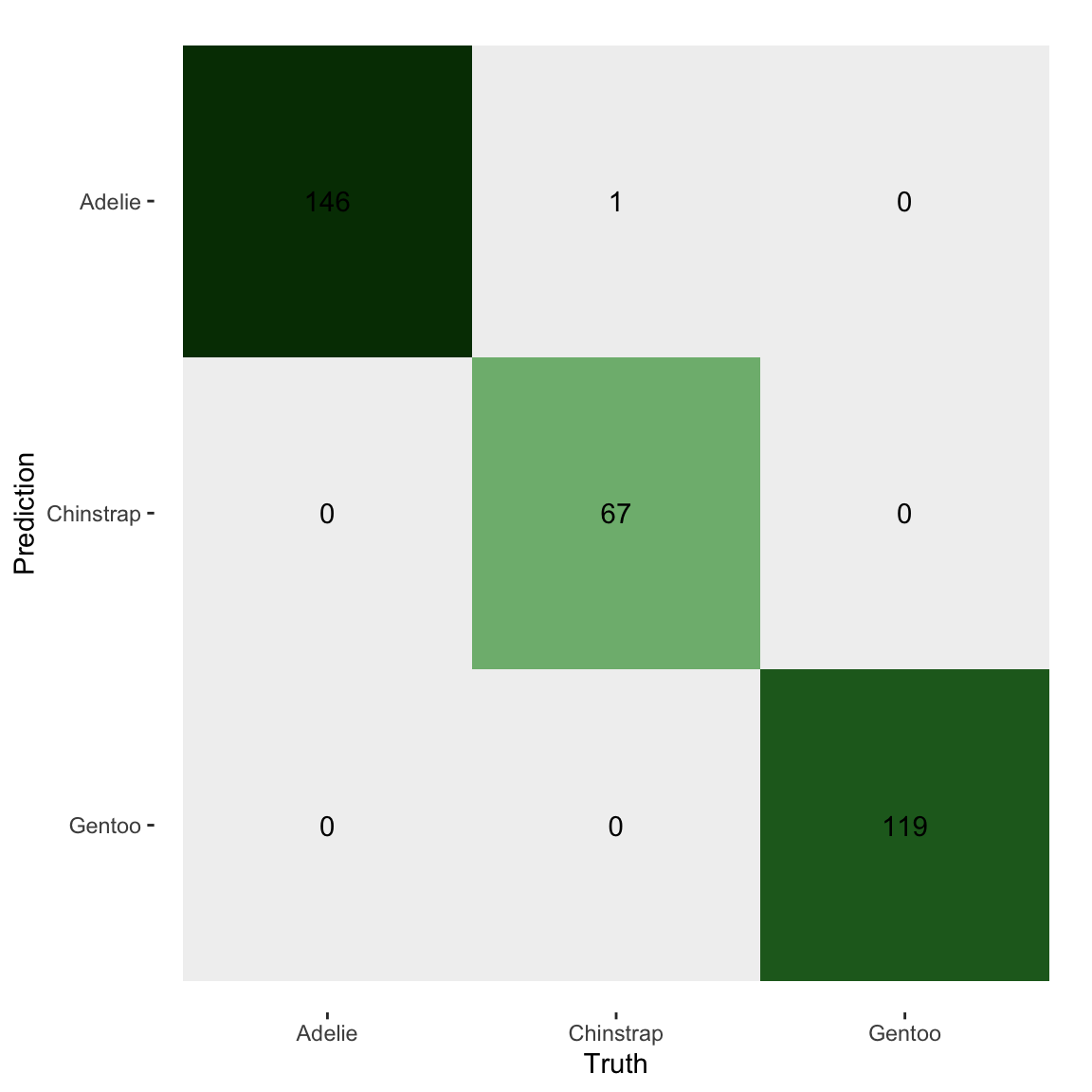
Avoid overfitting
?initial_split
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 bal_accuracy macro 0.987last_fit fits on the training split and evaluates on the test split# A tibble: 1 × 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 bal_accuracy macro 0.987 Preprocessor1_Model1